Proteins: Structure, Function, and Bioinformatics (2023). DOI: 10.1002/prot.26496”>
Credit: Proteins: Structure, Function, and Bioinformatics (2023). DOI: 10.1002/prot.26496
During the COVID-19 pandemic, Professors Joel Sussman and Israel Silman found themselves mentoring Chinese students online. What transpired from this experience was unexpected—a groundbreaking research endeavor on protein evolution that has the potential to reshape our comprehension of how new proteins emerge.
Initially hesitant due to the virtual nature of the interaction with undergraduate students from prominent Chinese universities, Sussman reflects on his skepticism. However, both Sussman and Silman, renowned for their collaborative studies on protein structure and function at the Weizmann Institute of Science, decided to conduct tutorials for a team of four students. This initiative was part of the YutChun-Weizmann Program under the leadership of Prof. Binghai Yan.
Breaking from the traditional Chinese academic norms, Sussman and Silman encouraged the students to address them by their first names and fostered an environment conducive to critical thinking. What started as a request for a review of an old paper on protein sequence variations led to a remarkable outcome. The students provided a comprehensive critique, offering a contemporary perspective and proposing revisions to certain conclusions using innovative methodologies.
Jing Liu, one of the mentees, highlighted the stark contrast in academic culture between China and her experience at the Guangdong campus of the Technion—Israel Institute of Technology. She emphasized the significance of having a supportive supervisor who valued open discussions—a rarity in many Chinese universities.
The online tutorials evolved into engaging discussions, with a particular focus on a 2017 study by Czech scientists that hinted at a paradigm shift in the understanding of protein evolution.
Challenging the Established Norms
The prevailing belief among scientists was that existing proteins evolved through refinement, and the emergence of entirely new proteins was a rare occurrence. However, recent revelations have challenged this notion. Genomic comparisons across various species unveiled genes encoding “newly born” proteins, suggesting a continuous generation of novel proteins originating from noncoding regions of the genome.
The Czech study, which captivated Liu and her mentors, introduced a new dimension to this discourse. By creating hypothetical protein sequences through random reshuffling of existing genes and observing their folding patterns in the lab, the researchers demonstrated that a significant portion exhibited compact structures akin to natural proteins.
This unexpected discovery prompted Sussman to acknowledge the transformative implications, noting the implications of random protein sequences folding in such a manner.
Silman elaborated on the critical role of protein folding in biological processes, emphasizing that the ability of “never-born” proteins to fold suggested the potential for new proteins to fulfill essential biological functions.
Unveiling New Horizons
To delve deeper into the folding potential of “newly-born” proteins, Sussman, Silman, Liu, and Yuan embarked on a structural study aided by artificial intelligence (AI) tools. Leveraging cutting-edge algorithms capable of predicting protein structures based solely on amino acid sequences, the team navigated the challenge posed by orphan proteins—those lacking evolutionary predecessors.
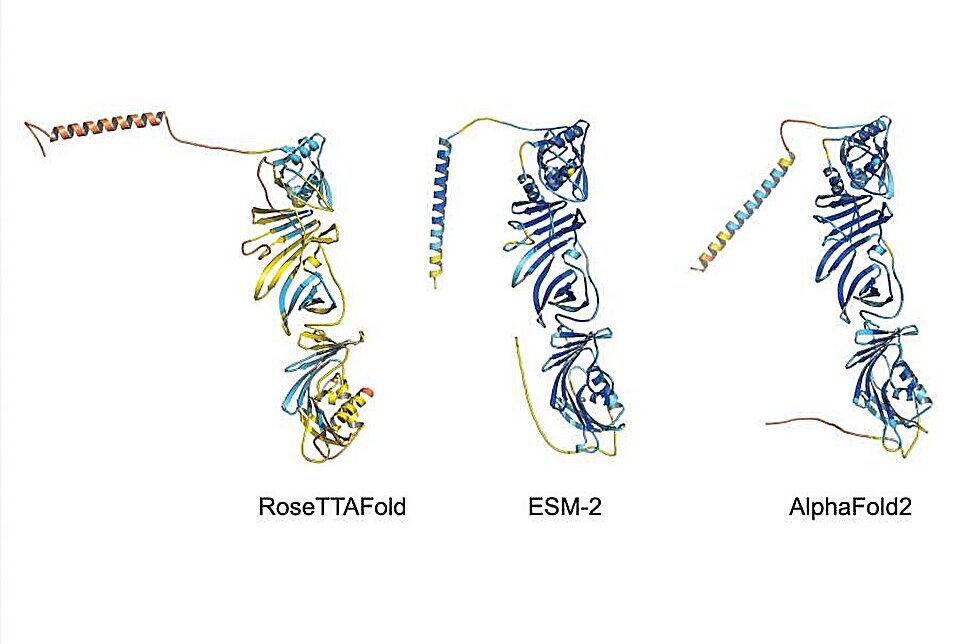
Employing AI algorithms such as AlphaFold2, RoseTTAFold, and ESMFold, the team successfully predicted the 3D structures of “never born” proteins from the Czech study. Subsequent application of these algorithms to “newly-born” orphan proteins revealed intriguing insights. Despite limited experimental characterization, the AI predictions indicated that some orphan proteins exhibited compact folding patterns, hinting at unexplored biological functions.
The team’s meticulous analysis, encompassing orphan proteins with known functions but undefined structures, uncovered novel folds previously unseen in protein databases. These unique structural features suggest untapped potential for diverse applications, ranging from environmental remediation to therapeutic interventions.
Sussman underscored the transformative nature of their research, challenging conventional evolutionary paradigms and opening new avenues for biochemical exploration. Silman echoed this sentiment, emphasizing the broader implications for understanding life’s origins and the emergence of novel biological traits.
Liu and Yuan, integral members of the research team, have since pursued further academic endeavors, underscoring the lasting impact of their collaborative efforts. The study, recently published in the journal Proteins: Structure, Function, and Bioinformatics, marks a significant milestone in unraveling the mysteries of orphan proteins and their biological significance.
The research conducted by Sussman, Silman, and their team not only sheds light on the evolutionary dynamics of proteins but also underscores the potential for AI-driven approaches to unveil novel biological insights. By exploring the structural and functional landscapes of orphan proteins, this study paves the way for future discoveries and underscores the boundless possibilities inherent in the realm of protein science.
More information:
Jing Liu et al, Do “Newly Born” orphan proteins resemble “Never Born” proteins? A study using three deep learning algorithms, Proteins: Structure, Function, and Bioinformatics (2023). DOI: 10.1002/prot.26496
Provided by
Weizmann Institute of Science
Citation:
Using three AI protein prediction tools, study uncovers new wrinkles in the folding story of ‘orphan’ proteins (2024, February 14)
retrieved 14 February 2024
from https://phys.org/news/2024-02-ai-protein-tools-uncovers-wrinkles.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no part may be reproduced without the written permission. The content is provided for information purposes only.