Researchers at the University of Toronto have employed an artificial intelligence framework to revamp a critical protein crucial in the administration of gene therapy.
The research, detailed in the publication Nature Machine Intelligence, outlines a fresh approach to enhancing proteins to alleviate immune reactions, thereby enhancing the effectiveness of gene therapy and diminishing side effects.
According to Michael Garton, an assistant professor at the Institute of Biomedical Engineering in the Faculty of Applied Science & Engineering, “Gene therapy shows great potential, but the body’s existing immune response to viral vectors significantly impedes its success. Our study focuses on hexons, a key protein in adenovirus vectors, which, if not for the immune challenge, possess immense promise for gene therapy.”
The immune responses provoked by serotype-specific antibodies present a significant barrier to guiding these vectors to their intended target, leading to decreased efficacy and severe adverse effects.
To tackle this challenge, Garton’s team utilized AI to tailor variants of hexons that diverge from natural sequences.
“We aim to create a design that differs from all human variants and consequently remains unidentifiable to the immune system,” explains Ph.D. candidate Suyue Lyu, the lead author of the research.
Conventional methods of protein design often entail extensive trial and error processes and escalating expenses. By leveraging an AI-driven strategy for protein design, researchers can introduce a higher level of diversity, reduce expenses, and swiftly generate simulation scenarios before focusing on specific targets for experimental evaluation.
Despite the existence of numerous protein-designing frameworks, crafting new variants can be daunting due to the limited availability of natural sequences and the considerable size of hexons—comprising an average of 983 amino acids.
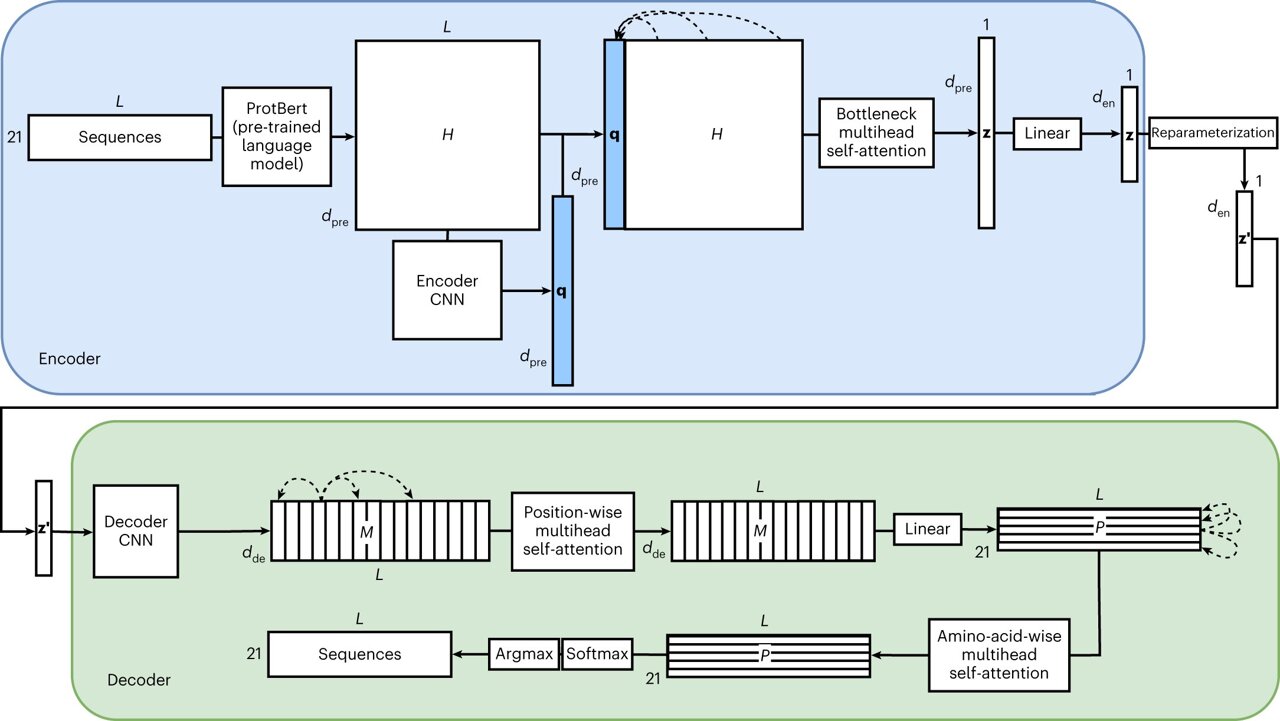
In light of this challenge, Lyu and Garton devised a distinct AI framework named ProteinVAE. This model can be trained to grasp the attributes of lengthy proteins with minimal data. Despite its streamlined design, ProteinVAE showcases a generative capacity comparable to more extensive models currently available.
“Our model leverages pre-trained protein language models for efficient learning on modest datasets. We also integrated several customized engineering techniques to render the model apt for generating lengthy proteins,” mentions Lyu, highlighting that ProteinVAE was intentionally crafted to be resource-efficient.
Lyu further elaborates, “In contrast to significantly larger models that necessitate substantial computational resources for lengthy protein design, ProteinVAE facilitates rapid training and inference on standard GPUs. This aspect could enhance the model’s accessibility for other academic laboratories. Our AI model, validated through molecular simulation, illustrates the capability to modify a notable portion of the protein’s surface, potentially circumventing immune reactions.”
The subsequent phase involves experimental validation in a laboratory setting, as outlined by Lyu.
Garton envisions that the AI model could find utility beyond gene therapy protein design and might feasibly be extended to support protein design for various disease scenarios.
“This study suggests that we may have the ability to craft new subspecies and even species of biological entities using generative AI,” he asserts, “and these entities offer therapeutic potential for innovative medical interventions.”