by Harvard University
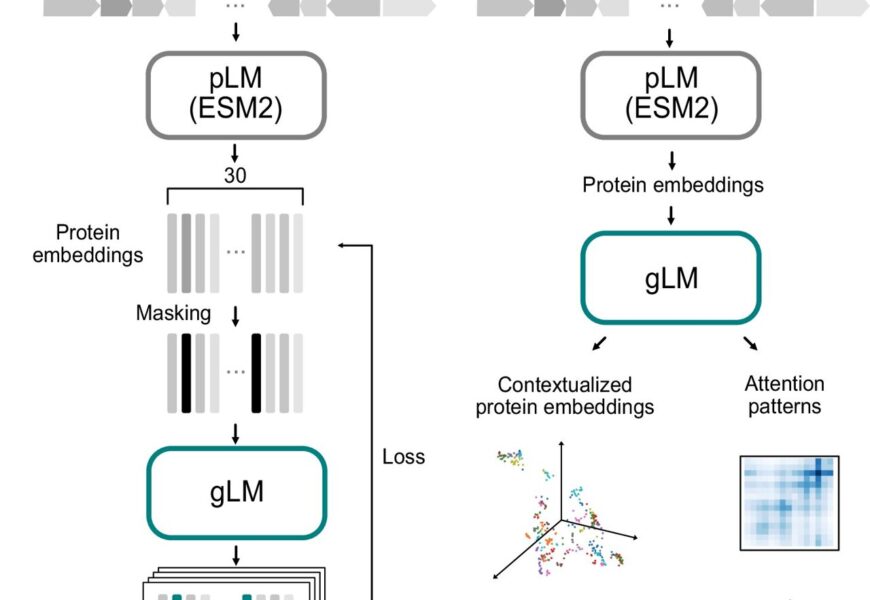
 gLM training and inference schematics. A During the training phase, contigs (which are contiguous genomic sequences) containing up to 30 genes undergo an initial translation into proteins. These proteins are then processed using a protein language model (pLM) encoder (ESM2). The generation of masked inputs involves random masking at a probability of 15%, following which a genomic language model (gLM)—a transformer encoder—is trained to make four predictions for each masked protein along with their associated likelihoods. The training process involves calculating the loss based on both the predictions and likelihoods. B When it comes to inference, inputs are created from a contig utilizing the output from ESM2. The contextualized protein embeddings (which are the hidden layers of gLM) and attention patterns play a crucial role in various downstream tasks. Credit: Nature Communications (2024). DOI: 10.1038/s41467-024-46947-9
gLM training and inference schematics. A During the training phase, contigs (which are contiguous genomic sequences) containing up to 30 genes undergo an initial translation into proteins. These proteins are then processed using a protein language model (pLM) encoder (ESM2). The generation of masked inputs involves random masking at a probability of 15%, following which a genomic language model (gLM)—a transformer encoder—is trained to make four predictions for each masked protein along with their associated likelihoods. The training process involves calculating the loss based on both the predictions and likelihoods. B When it comes to inference, inputs are created from a contig utilizing the output from ESM2. The contextualized protein embeddings (which are the hidden layers of gLM) and attention patterns play a crucial role in various downstream tasks. Credit: Nature Communications (2024). DOI: 10.1038/s41467-024-46947-9
Artificial intelligence (AI) systems such as ChatGPT have become ubiquitous across various domains, offering recommendations for entertainment choices and aiding in navigation. Can these AI systems extend their capabilities to understand the language of genetics and assist biologists in making groundbreaking scientific discoveries?
In a recent research publication in Nature Communications, a team of interdisciplinary researchers led by Yunha Hwang, a Ph.D. candidate in the Department of Organismic and Evolutionary Biology (OEB) at Harvard University, introduced an AI system capable of interpreting the complex language of genomics.
Genomic language serves as the fundamental code of biological systems, encapsulating the functional aspects and regulatory principles embedded within genomes. The researchers pondered the question, “Is it feasible to develop an AI engine that can ‘comprehend’ the genomic language, gaining fluency in understanding the functions and regulations of genes?” To address this, the team fed a vast microbial metagenomic dataset—the most extensive and diverse genomic dataset available—into the machine learning model to establish the Genomic Language Model (gLM).
Hwang remarked, “In the field of biology, researchers operate within the confines of a known vocabulary. However, this existing lexicon represents only a minute fraction of biological sequences.” She highlighted the challenge posed by the exponential growth of genomic data, which surpasses human processing capacity.
Large language models (LLMs) like GPT4 acquire word meanings by processing extensive and varied textual data, enabling them to grasp semantic relationships. Similarly, the Genomic Language Model (gLM) learns from a wide range of metagenomic data derived from microbes residing in diverse habitats such as the ocean, soil, and the human gut.
Through this data, gLM gains insights into the functional “semantics” and regulatory “syntax” of individual genes by discerning the associations between genes and their genomic context. Like LLMs, gLM functions as a self-supervised model, extracting meaningful gene representations solely from data without the need for human-assigned annotations.
While scientists have sequenced genomes of extensively studied organisms like humans, E. coli, and fruit flies, a significant portion of genes within these genomes remains poorly characterized.
Professor Peter Girguis, a senior author from the OEB department at Harvard, reflected on the limitations of traditional biological research methodologies, emphasizing the sequential nature of gene analysis that often leads to discoveries within known boundaries. In contrast, gLM empowers biologists to explore the context surrounding unknown genes, shedding light on their roles within similar gene clusters. By identifying groups of genes that collaborate towards a common goal, the model provides insights that transcend conventional annotations.
Hwang elucidated, “Genes, akin to words, exhibit varying ‘meanings’ based on their contextual placement. Conversely, genes with distinct sequences can serve ‘synonymous’ functions. gLM introduces a nuanced framework for comprehending gene functionality, departing from the conventional one-to-one mapping approach, which fails to capture the dynamic and context-specific nature of genomic language.”
The study showcases gLM’s ability to predict enzymatic functions, co-regulated gene clusters (referred to as operons), and offer genomic insights that aid in predicting gene functions. Additionally, the model discerns taxonomic details and context-specific dependencies related to gene functions.
Remarkably, gLM operates without prior knowledge of the specific enzymes or bacterial origins within the sequences. Leveraging its exposure to numerous sequences and understanding of their evolutionary relationships during training, gLM deduces functional and evolutionary links between sequences.
Martin Steinegger, an expert in bioinformatics and machine learning at Seoul National University, commended gLM for integrating gene neighborhoods with language models, thereby enhancing the understanding of protein interactions on a broader scale.
By employing genomic language modeling, biologists can unearth novel genomic patterns and unravel undiscovered biological phenomena. gLM stands as a significant milestone in collaborative interdisciplinary efforts propelling advancements in life sciences.
Hwang expressed optimism about gLM’s potential to uncover hidden patterns within poorly annotated genomes, guide experimental validations, and facilitate the discovery of novel biological functions and mechanisms that could address challenges related to climate change and bioeconomy.
More information:
Yunha Hwang et al, Genomic language model predicts protein co-regulation and function, Nature Communications (2024). DOI: 10.1038/s41467-024-46947-9
Provided by Harvard University
Citation:
Deciphering genomic language: New AI system unlocks biology’s source code (2024, April 3) retrieved 3 April 2024 from https://phys.org/news/2024-04-deciphering-genomic-language-ai-biology.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no part may be reproduced without the written permission. The content is provided for information purposes only.